Screening and activity study of human scFv antibody targeting the extracellular domain of FGFR3
-Molecular Devices
Fibroblast growth factor (FGF) is the largest family of mesodermal and epithelial cell growth and differentiation polypeptide factors. FGFs play important roles in many biological processes, such as embryonic development, wound healing, hematopoiesis, and angiogenesis. In addition, studies have shown that FGFs can increase the invasiveness of a variety of tumor cells, such as prostate, bladder, kidney, breast, pancreas and other tumors.
At present, more than 20 kinds of FGFs have been found, which have different effects on different types of cells. However, only five FGF receptors (FGFR) were found. At the protein level, these receptors have between 55% and 72% homology. The FGFR structure includes an extracellular ligand binding region, a transmembrane region, and an intracellular kinase region. The ligand binding region comprises three different immunoglobulin-like domains (referred to as immunoglobulins I, II and III). Different splicing effects of FGFR1-3 mRNA form two subtypes α and β. Among them, FGFR3 has two different mutants IIIb and IIIc. These two variants have different affinity activities: IIIc is more widely distributed and can bind to various FGFs (FGF1, FGF2, FGF4, and FGF9); IIIb preferentially binds to FGF1 and can bind to FGF8 and FGF9 to a lesser extent. In the presence of heparin, FGFs induce receptor dimerization following binding of FGFs to FGFRs, causing autophosphorylation of the intracellular kinase domain and activation of downstream signaling cascades. After ligand receptor binding, FGFs initiate a variety of signal transduction pathways: elevated intracellular calcium levels, induction of mitogen-activated protein kinases and protein kinase C pathways, activation of adenylate cyclase, and induction of proto-oncogenes C-myc and c- fos.
It has been found that FGFR3 undergoes specific mutations that cause its tyrosine kinase activity to activate, causing some syndromes associated with skeletal development, multiple myeloma, neck tumors, and bladder tumors. Recent studies have found that FGFR signaling is essential for prostate tumor cells to survive in vitro. Recently, FGFR3 has been used as a potential therapeutic target for multiple myeloma. Although there is indeed evidence that activated FGFR3 mutations are present in tumor tissues, little is known about the expression of FGFR3 in tumor tissues. Recently, after gene expression analysis using gene chip technology, it was found that FGFR3 was overexpressed in bladder tumor samples compared to normal tissues. Gene expression levels were further confirmed at the protein level by Western blot and immunohistochemical analysis. In fact, this overproduction of protein appears to be more likely to occur in transitional tumors than genetic mutations. All of these data suggest that FGFR3 may be a very attractive therapeutic target for urological tumors. Bladder tumors are one of the second most common malignancies in the genitourinary system. Approximately 40%-50% of bladder tumors exhibit mutations in the FGFR3 gene; the probability of epidermal tumorigenesis (80%) is greater than that of invasive tumors.
With the growing interest in FGFR3 as a therapeutic target for different tumors, and the recent discovery of its overexpression in transitional cell tumors, we have begun to develop human antibodies for therapeutic use using phage display technology. Phage antibody display is currently the best method for developing human antibodies for research, clinical, and therapeutic use. However, for antibody development, the FGFR3 molecule is very difficult to honed because the homology of FGFR3 in mice and humans is very high (92%). Only recently, it has been reported that a Fab fragment for one FGFR3 subtype was developed by using a very large (2.1*1010) commercial Fab library. In our experiments, we used two published scFv antibody libraries, Tomlinson I + J (MRC Geneservices, Cambridge, United Kingdom). The storage capacity of both libraries is about 1.4*108. Compared with IgG and Fabs, scFvs has better tumor infiltration, can be cleared more quickly, and has better specificity. In this report, we have screened for human scFv antibodies specific for subtype IIIc of FGFR3a. These antibodies have been shown to react with the bladder tumor cell line RT112 by FACS and inhibit cell proliferation with potential for further therapeutic use.
Materials and Method
Cell lines, proteins, antibodies:
RT112, HEK293; recombinant human FGFR3a (IIIc)/Fc, FGFR1a (IIIc)/Fc, FGF9, FGF1 and epidermal growth factor. Mouse anti-human FGFR3 monoclonal antibody, goat anti-human IgG (Fc specific), mouse anti-c-myc monoclonal antibody, tubulin inhibitor; HRP-anti-c-myc antibody, anti-6His antibody, anti-M13 antibody, HRP - anti-M13 monoclonal antibody, FITC-rabbit anti-mouse IgG, R-phycoerythrin goat anti-mouse IgG, murine IgG TrueBlot.
FGFR3 cDNA Clone and Cell Transfection
The extracellular region of FGFR3 was fused to the Fc C-terminus of human IgG, and the expression vectors pcDNA3.1-FGFR3(IIIc) WT-Fc and pcDNA3.1-FGFR3(IIIc)S249C-Fc were constructed. Then, HEK293T cells were transfected. Protein expression and activity were detected by immunoblotting and FACS.
Screening of FGFR3-specific scFv antibodies from phage libraries
Human scFv phage library Tomlin-son I + J, helper phage KM13, E. coli TG1 and HB2151. Two phage libraries were separately cultured, and then 1:1 mixed phage were used for screening. Phage library screening and scFv expression were performed as shown in Scheme 1. For the first round of screening, microplates were coated with 1 ug of FGFR3 or human IgG protein. Phage were first incubated with human IgG for 1 hour to remove phage that could react with Fc; and incubated with FGFR3-coated wells for 2 hours. Finally, the microplates were washed 10 times with 0.1% PBST (20 washes in the next round of screening) and the microplates were treated with 100 ul trypsin to elute the bound phage. The phage obtained by elution was subjected to another round of panning according to the method described by Goletz et al.
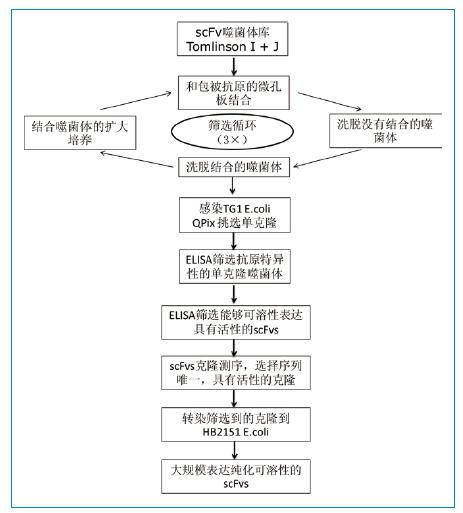
Figure 1: Flow chart of phage library screening FGFR3-specific scFv antibody
Phage ELISA
The microporous plates were coated with 0.3 ug of FGFR3, FGFR1 or human IgG protein, washed, blocked, and phage suspensions purified and screened at different concentrations were added. After the incubation, HRP-anti-M13 antibody was added, the substrate was added for color development, and the result was read at 450 nm.
Monoclonal phage ELISA
Phage harvested after two or three rounds of screening were grown to clone, and then picked up into 96-well plates containing 100 ul of 2TY medium using a QPix high-throughput automated cloning screening system (Molecular Devices), 37 degrees The culture was carried out on the next day, and the culture was diluted 1:100 with the same medium, followed by shaking culture at 37 °C for 2 hours, and 25 ul of 2TY medium containing 109 helper phage KM13 was added, and the culture was continued for 1 hour. The cells were centrifuged, resuspended in 2TY medium, and cultured at 30 degrees overnight. Finally, centrifugation was carried out, and 50 ul of the supernatant was taken for monoclonal phage ELISA analysis.
Expression and purification of soluble scFv antibody and detection by ELISA
The highly specific clone obtained was induced by E. coli HB2151, the cells were harvested by centrifugation, the bacterial periplasmic cavity protein was extracted, and finally purified by affinity chromatography. The purified fractions were analyzed by SDS-PAGE and Coomassie blue staining. The purified fraction is further separated from the monomer of the scFv protein by molecular sieve chromatography (scFv is prone to dimer or multimer).
Surface plasmon resonance analysis
Binding kinetics of soluble scFv and FGFR3 were analyzed using BiacoreX and Kd values ​​for each purified scFv were calculated; specific binding sites for each scFv were determined using competitive binding assays. Using the same method to analyze whether scFvs and FGF9, FGF1 and EGF can compete for binding to FGFR3.
Flow cytometry and confocal microscopy analysis
The phage scFv and purified soluble scFv were analyzed for binding activity to FGFR3 at the cellular level using a cytometer. The antibody binding activity was further analyzed by treating RT112 cells and then using confocal microscopy.
Cell proliferation analysis
RT-112 cells were treated with different concentrations of anti-FGFR3 scFvs antibody (0.02-2 umol/L) and after 48 hours, cells were stained with MTT and then read at 570 nm. Cell viability was calculated by the following formula: Abs-scFv treated cells/Abs-control cells.
result
Screening of FGFR3-specific scFv antibodies: As shown in Table 1, a total of 288 clones were randomly selected and 15 FGFR3-specific antibody clones were obtained by ELISA. It was found by sequencing that the sequence of 6 clones in 15 clones was unique. The 3A and 1D sequences have a stop codon inside and only have a base mutation in the CDR2 region of the VL. The sequences of the other four clones are quite different, and the main differences are concentrated in CDR2 and CDR3. Among them, CDR3 is highly variable in both VH and VL, and mutations in CDR2 are mainly concentrated in VH.
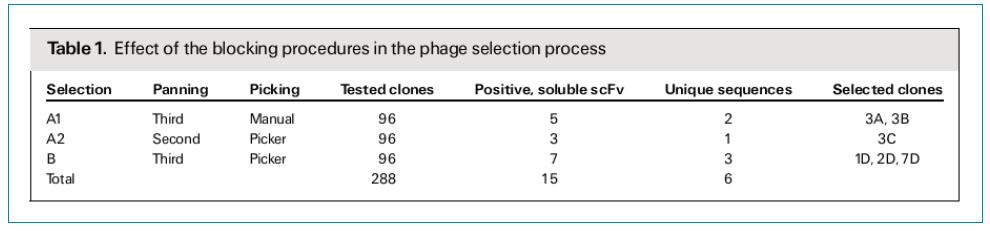
ELISA-specific identification revealed that, as shown in Figure 2, although FGFR1 and FGFR3 are highly homologous (>62%), the six selected scFv clones have significant binding specificity for FGFR3; instead, binding activity to FGFR1 Very weak, similar to the reaction of the negative control human IgG.
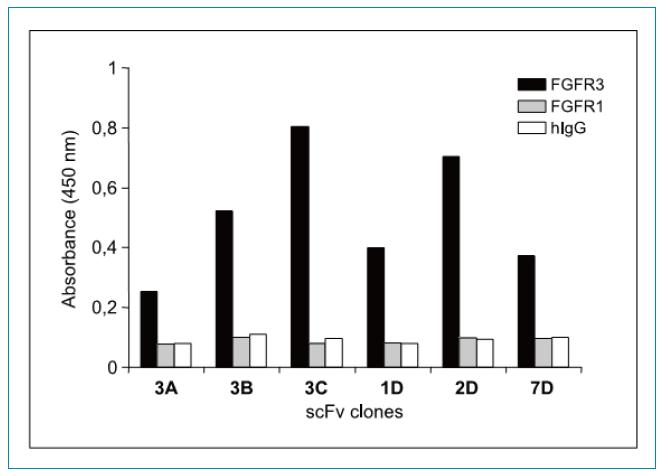
Figure 2: ELISA assay for selected soluble scFvs binding specificity. The scFv-pIII fusion protein obtained from the culture supernatant was assayed for binding activity to FGFR3-Fc (black), FGFR1-Fc (grey), and negative control human IgG (white) by an ELISA method.
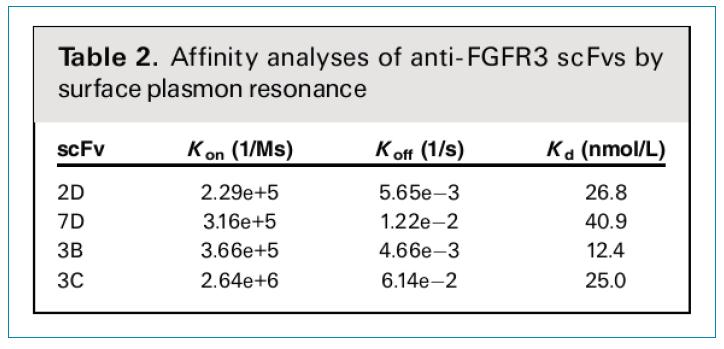
The affinity of the purified monomeric scFv antibody and FGFR3 was analyzed using Biacore. As shown in Table 2, the Kon, Koff, and Kd values ​​of different scFvs were examined. The Kd value varied between 40.9 and 12.4 nmol/L, indicating that the screened scFv has moderate to high affinity activity. If you consider that the phage used is only 1.6*108, which is a medium size storage, this result is very good.
Activity of scFv antibody to FGFR3 expressed by RT112 cells: The binding activity of scFvs and native FGFR3 protein expressed by bladder cancer RT112 cells was examined using FACS. As shown in Figure 3A, six scFvs phage clones were first detected, and all six clones were able to react significantly with bladder cancer RT112 cells. Then, the binding activity of the purified four scFvs proteins (3B, 3C, 2D, 7D) was examined, and it was found that 3C and 7D can react remarkably with RT112 cells, and the reactivity of 2D and 3B is relatively weak. 3C and 7D stained cells were observed using confocal microscopy, further showing the membrane protein staining characteristics of scFv (Fig. 3A).
The affinity specificity of scFv and FGFR3 was further demonstrated by immunoprecipitation methods under non-denaturing conditions. As shown in Figure 3B, both scFvs (3C and 7D) after purification produced significant protein conditions at the correct position, further demonstrating that the two scFv antibodies recognize the soluble and native FGFR3 receptors.
Reactivity of scFv antibody and FGFR3 mutant: Since FGFR3 is frequently mutated in bladder cancer cells, binding activity of two scFv antibodies and FGFR3 S249C mutant was also examined, and FGFR3 S249C has been reported in bladder cancer cells. The most frequent form of mutation. The above mutation was introduced into FGFR3 by PCR and the mutation site was confirmed by DNA sequencing (Fig. 4A). The transfected HEK293T cells were verified by western blot to correctly express the mutated FGFR3S249C protein (Fig. 4B), and FGFR3S249C has a similar expression level as wild-type FGFR3WT. FACS analysis demonstrated that both scFvs 3C and 7D recognized HEK293T cells expressing the mutant FGFR3 protein and were even more reactive than cells expressing the wild-type FGFR3 protein (Fig. 4C).
Inhibition of RT112 cell proliferation: We examined the effectiveness of scFvs in inhibiting RT112 tumor cell proliferation in vitro. Three different concentrations of scFv were added to the RT112 cells. As shown in Figure 5A, both scFv 3C and 7D were able to significantly inhibit cell proliferation, and the inhibitory effect was positively correlated with the concentration of antibody added. At the concentration of 2 umol/L, the inhibitory activity was the strongest, but at 0.2 umol/L, there was still significant inhibitory activity.
In addition, the effects of different growth factors on the inhibitory effect of scFvs were examined. As shown in Figure 5B, the inhibitory effect of scFvs has protein factor specificity. Both 3C and 7D can inhibit proliferation induced by FGF9, but only 7D can inhibit FGF1-induced proliferation. Neither of the two scFvs antibodies inhibited EGF-induced cell proliferation. In addition, the addition of soluble FGFR3 protein masked the inhibitory effect of scFv. Morphologically, after 3C and 7D treatment, cell morphology changes, especially after 7D treatment, which can cause cell vacuol formation (Fig. 5C).
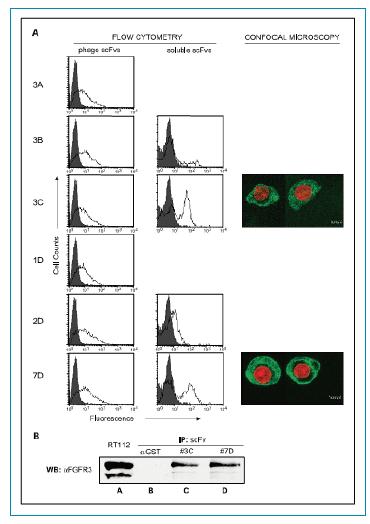
Figure 3. FACS and confocal microscopy analysis of the binding activity of phage, soluble scFvs protein and RT112 bladder cancer cells. A. The FACS histogram shows the binding activity (thick line) and background fluorescence (gray) of each scFv clone. RT112 cells were labeled with 3C and 7D scFvs and cells were stained with FITC fluorescent antibody and DAPI. Imaging under a laser confocal microscope revealed that the (green) scFvs protein binds to the cell membrane and the (red) DAPI stains the nuclear region. B. RT112 cell lysate FGFR3 IP analysis. A. RT112 cell lysate; B. Negative control; C, scFv3C; D, scFv7D.
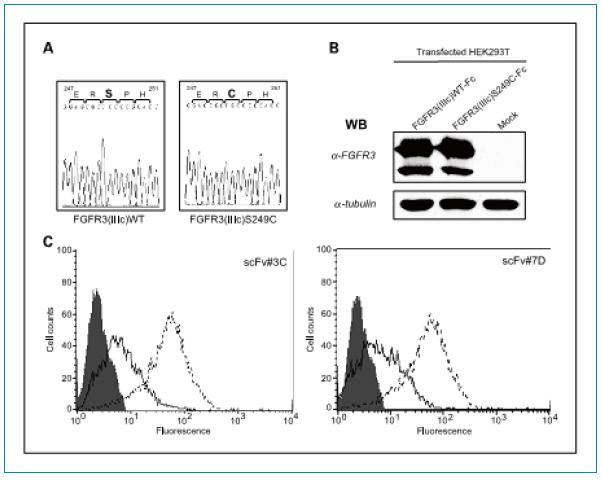
Figure 4. Reactivity of screened scFvs and FGFR3 mutant extracellular regions. A. Sequencing revealed a mutation introduced into the DNA sequence: Ser(TCC) 249 was mutated to Cys (TGC). B. Immunoblot analysis demonstrated expression of wild-type FGFR3 and mutant FGFR3 (S249C) in HEK293T cells. C. FACS analysis of the binding activity of the scFv protein and expression of wild-type FGFR3 and mutant FGFR3 (S249C) HEK293T cells. In the histogram: gray indicates a negative control; black line indicates FGFR3 (IIIc) WT; and broken line indicates FGFR3 (IIIc) S249C.
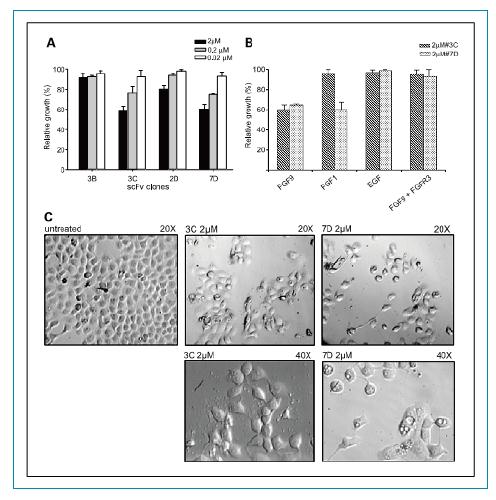
Figure 5. Inhibition of proliferation of RT112 cells by FGFR3-specific scfvs antibodies. A. Inhibitory activity of different concentrations of 3B, 3C, 2D and 7D scFvs antibodies on RT112 cells. Analysis of cell number and morphology after C. 2 umol/L 3C and 7DscFvs treatment of cells.
discuss
FGFR3 is a receptor tyrosine kinase that plays a role in cell growth and tumor formation by activating different signaling pathways. In addition to bladder cancer, FGFR3 is mutated and overexpressed in many types of tumors, so FGFR3 can be an excellent and effective therapeutic target for tumors. Previous studies have shown that the extracellular ligand binding domain of soluble native FGFR3 has the effect of inhibiting cell proliferation by competing to bind ligands and reduce ligand and cell surface receptor binding. Antibody fragments may mimic similar mechanisms to inhibit cell proliferation. In fact, many studies have shown that the mechanism by which 3C and 7D can inhibit the proliferation of RT112 cells is that antibody fragments directly block the interaction of FGF-FGFR3, thereby inhibiting the activation of FGFR3. However, the dissociation speed of 3C and 7D combined with FGFR3 is too fast (ie, the Koff value is too high).
1.22*10-2 and 6.14*10-2), a decrease in Koff means an increase in the Kd value, ie an increase in antibody affinity. Therefore, the sequence of 3C and 7D scFv can be used as a template, and on this basis, affinity maturation can be performed by CDR region mutation or somatic mutation simulation.
Reference material
1. Jorge MartÃnez-Torrecuadrada, Gabriela Cifuentes, Paula López-Serra, et al., Targeting the Extracellular Domain of Fibroblast Growth Factor Receptor 3 with Human Single-Chain Fv Antibodies Inhibits Bladder Carcinoma Cell Line Proliferation. Clin Cancer Res 2005;11: 6280-6290.
Toilet Cleaner,Toilet Bowl Cleaner,Toilet Cleaning,Toilet Cleaning Powder
Wuxi Keni Daily Cosmetics Co.,Ltd , https://www.kenidailycosmetics.com